Translational Research Integration & Support
The Translational Research Integration & Support platform creates and delivers end-to-end solutions that supports multi-institutional translational and pre-clinical studies, from study design to outcomes. We bring together expertise from cancer research institutes and academic centres across Singapore, with the common goal of developing, testing and validating potential new biomarkers and molecular diagnostics. Our platform boasts a comprehensive facilitation pipeline for translational and pre-clinical research, and functions as a coordinating hub for molecular assays and ex vivo tumour models. This supports our mission to promote collaboration and cohesiveness among academia and industry players in the cancer research community.
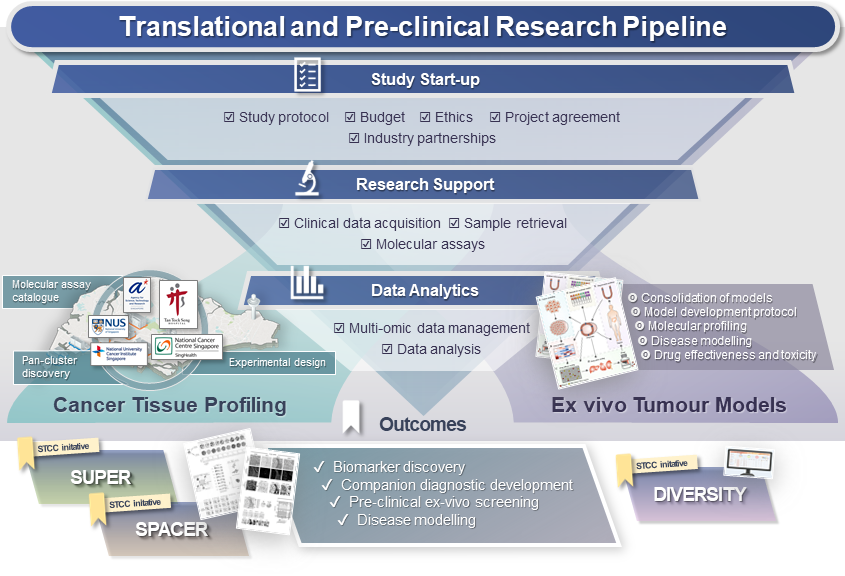
Translational and Pre-clinical Research Pipeline
A versatile end-to-end pipeline designed to enable and expedite multipartite translational and pre-clinical studies for our research partners, with STCC as the central point-of contact. We have established streamlined workflows and capabilities that synergise with multiple local institutions to support study initiation (preparation of study design and protocol, drafting of budget, ethics and multi-party agreements), case selection and sample acquisition, multi-omic molecular profiling at partner institutes, compound screening, as well as data analytics and data management.
Key focus:
· Cancer Tissue Profiling
Coordinating hub for non-interventional clinical studies and downstream translational research of clinical trials. We support all stages of these translational studies, from case identification, sample retrieval, molecular profiling (genomics, transcriptomics and spatial profiling) at multiple institutes to data analysis and management. Applications of the Cancer Tissue Profiling arm includes characterising therapeutic response / disease progression, patient stratification and biomarker development.
In our Molecular Assay Catalogue, we have compiled a list of local translational research capabilities and core facilities at partner cancer research institutes in Singapore. These assays are accessible to the Singapore cancer research community.
· Ex vivo Tumour Models
One-stop-shop to facilitate multi-institutional research involving ex vivo tumour models, such as identifying / validating biomarkers and promising drug candidates.
We will be setting up the DIVERSITY Tumour Model Virtual Catalogue (go-live in 2025): Curated collection of tumour organoid models of prevalent Asian cancers, with molecular and clinical annotations, generated by our research partners with ex vivo tumour model development expertise. We advocate and will facilitate the sharing of model development protocols to generate complex organoid models for research.
Key Outcomes
✓ Biomarker discovery
✓ Companion diagnostic development
✓ Pre-clinical ex-vivo screening of drug efficacy and toxicity
✓ Disease modelling
Example studies utilising this pipeline:
a) STCC Unified PD1/PDL1 Evaluation of Response (SUPER): This multi-institutional study aims to identify and profile matched pairs of immuno-oncology exceptional responders and hyper-progressors using a suite of nucleic acid-based and image-based assay platforms. Tissues samples are retrospectively retrieved from various hospitals and molecular profiling assays are performed at multiple institutes under STCC’s auspice. Data generated from this study will be used to validate an optimal combination of assays that best models the response to anti-PD1/PDL1 immunotherapy. A secondary objective is to identify and utilise new biomarkers to guide treatment choices for anti-PD1/PDL1 immunotherapy across multiple cancers
b) Spatial Phenotypes Ascribed to Cancers with Extreme Resistance (SPACER): This multi-institutional study aims to use advanced spatial phenotyping tools to investigate how the transcriptomic and proteomic landscapes in the tumour microenvironment (TME) contribute to or correlate with extreme resistance in four cancer cohorts (soft tissue sarcomas, breast cancer, multiple myeloma, hepatocellular carcinoma). Various spatial profiling platforms will be employed to identify molecular features in the TME that associate with primary resistance. The collected multidimensional spatial profiling data will be utilised to generate novel bioinformatics pipelines to define spatial phenotypes and biomarkers relevant to predicting outcomes in each of these cohorts.
For further inquiries, partnerships and projects, please fill in the online form.

